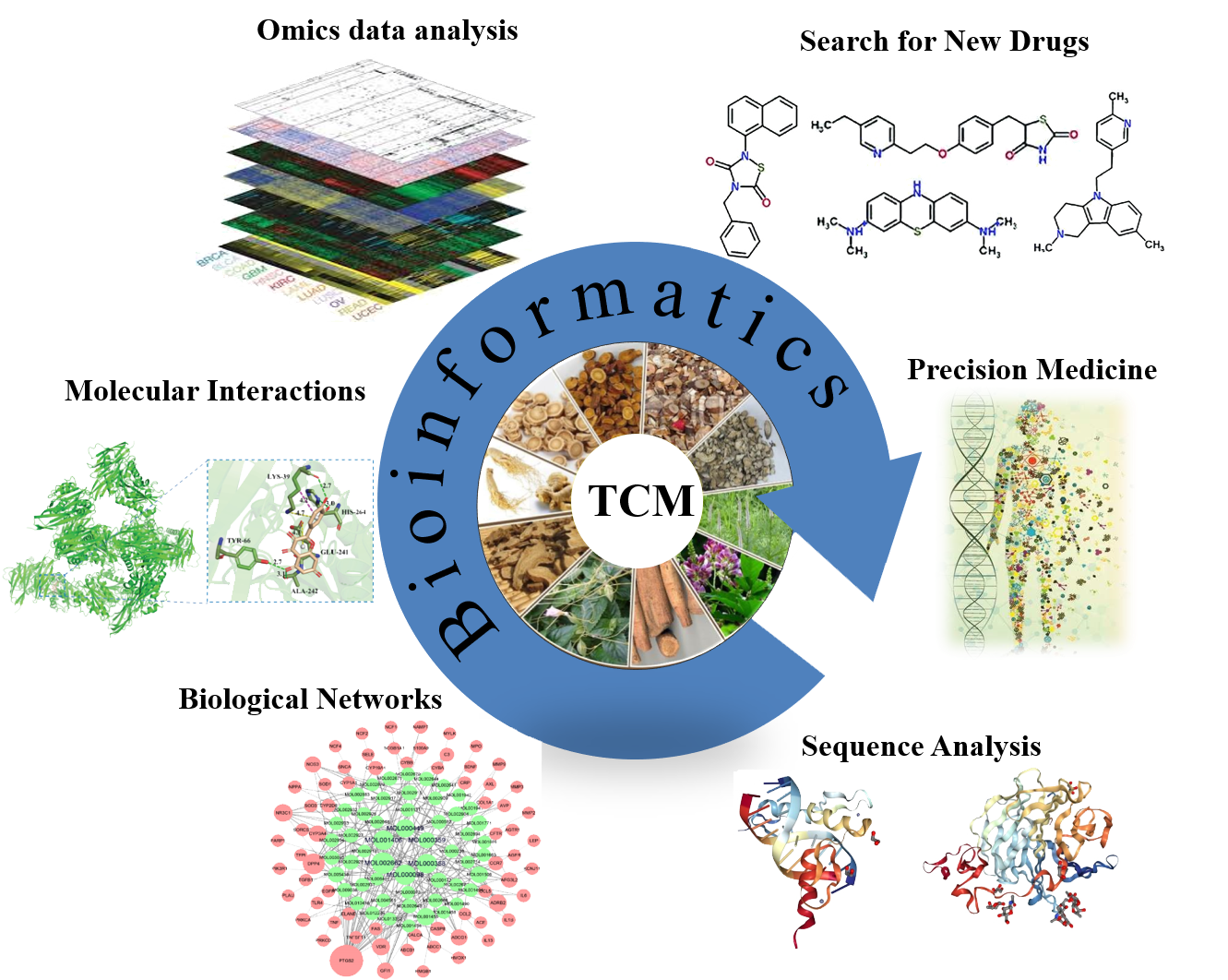
Databases
TCMPG: TCM Plant Genome Database
INPUT: An Intelligent Network Pharmacology Platform Unique for Traditional Chinese Medicine
KNIndex: a comprehensive database of physicochemical properties for k-tuple nucleotides
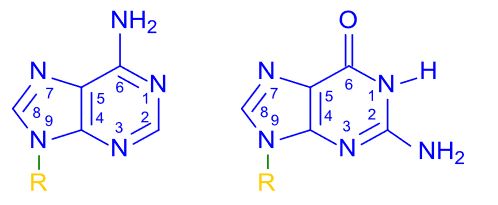 |
KNIndex is the abbreviation of K-Nucleotides Index database. The database deposits the original and standardized values of the physicochemical properties for k-tuple nucleotides (k = 1, 2, 3). |
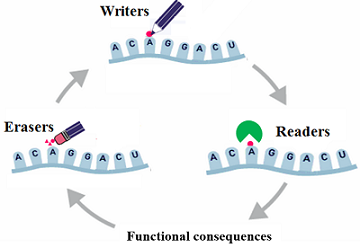
RNAWRE: a resource of writers, readers and erasers of RNA modifications
Pro54DB: a database for experimentally verified sigma-54 promoters
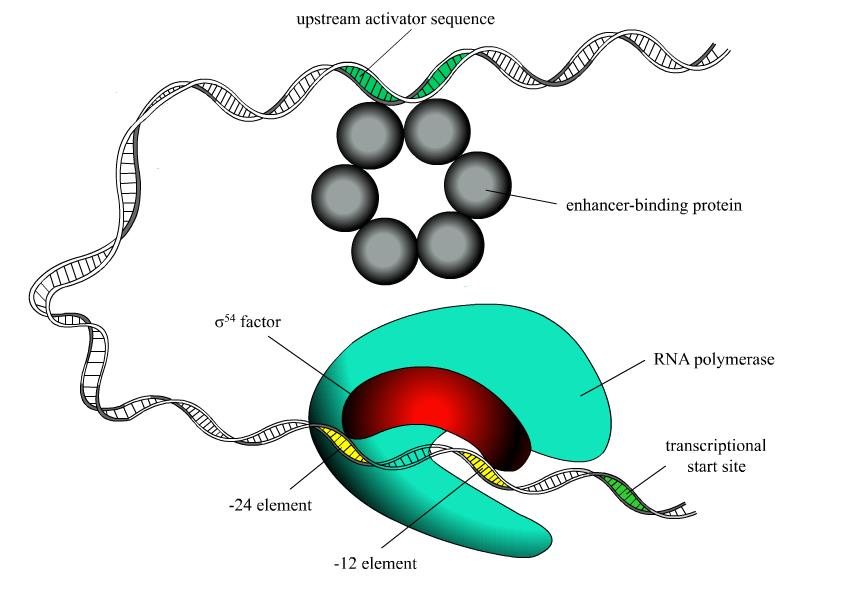 |
Pro54DB is a primary database on σ54 promoters and transcriptional regulation information curated from original scientific publications. |
AOD: a database for the antioxidant protein