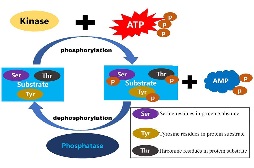
Protein
iPhoPred
:Predicting the Phosphorylation Sites in Human Protein
HBPred2.0
:Identification of hormone-binding protein
ApoliPred
:Identification of apolipoproteins
C2Pred
:Prediction of cell-penetrating peptides
IGPred
:Identification of immunoglobulins
iACP
: Predicting anticancer peptide
AodPred
: Predicting the antioxidant proteins
CaLecPred
: Predicting the cancerlectins
JPred
: Predicting the types of J-proteins
 |
The web-server was developed to identify the types of J-proteins by using reduced amino acid alphabet. The overall accuracy was over 94% by using jackknife cross-validation. |
subGolgi
: Prediction of subGolgi locations of proteins
TetraMito
: Prediction of submitochondria locations of proteins
ChloPred
: Prediction of subchloroplast locations of proteins
AcalPred
: Discriminating acidic enzymes from alkaline enzymes
ThermoPred
: Identification of thermophilic proteins
iVKC-OTC
: Identifying the subfamilies of voltage-gated potassium channe
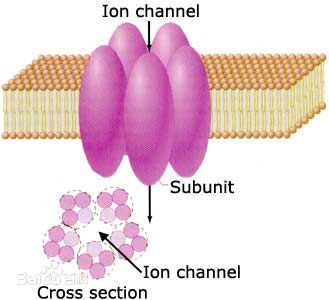
iCTX-Type
: Predicting the types of conotoxins in targeting ion channels
 |
The web-server iCTX-Type was developed to predict the types of conotoxins in targeting ion channels. The overall accuracy of 91.07% was achieved by using jackknife cross-validation. |
IonchanPred 2.0
: Identification of ion channels and their types
Lypred
: Prediction of bacterial cell wall lyase
PHYPred
: Prediction of bacteriophage enzymes and hydrolases
PHPred (version1.0)
and
PHPred (version2.0)
: Prediction of bacteriophage proteins located in host cell
PVPred
: Identification of phage virion proteins
Mycosub: Predicting subcellular localization of mycobacterial proteins
MycoSec: Identification of mycobacterial secretory proteins
MycoMemSVM: Identification of mycobacterial membrane proteins and their types