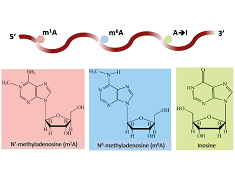
RNA
XG-PseU:an eXtreme Gradient Boosting based method for identifying pseudouridine sites
iMRM:is a predictor for identifying post-transcriptional modification sites
iRNA-Methyl
,
m6Apred
,
MethyRNA
,
M6ATH
and
iRNA(m6A)-PseDNC
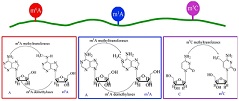
Identification of M6A sites in RNAs
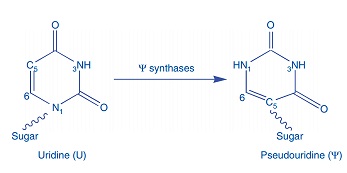
iRNA-PseU: Identifying RNA pseudouridine sites
PAI
and
iRNA-AI
: Predicting adenosine to inosine editing sites
 |
The web-server was developed to identify the Adenosine to Inosine editing sites in the D. melanogaster transcriptome. |
iRNA-PseColl
: Identifying m1A, m6A, and m5C sites in human
iRNA-3typeA
: Identifying m1A, m6A, and A-to-I sites in human and mouse
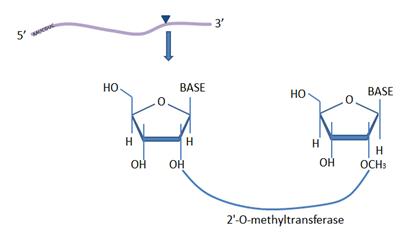
iRNA-2OM
: Identifying 2'-O-Methylation Sites in human
iRNAD
: identifying D modification sites
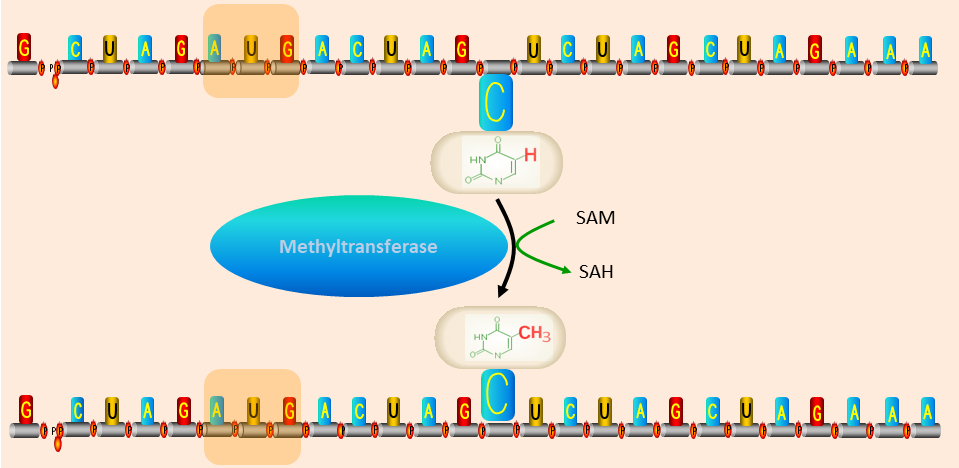
iRNA-m5C
: identifying m5C modification sites
iRNA-m7G
: identifying m7G modification sites
iRNA-m2G
: identifying m2G modification sites
iRNA-m6A
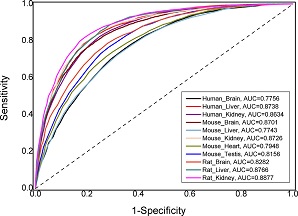
: identifying m6A modification sites in multiple tissues of mammals
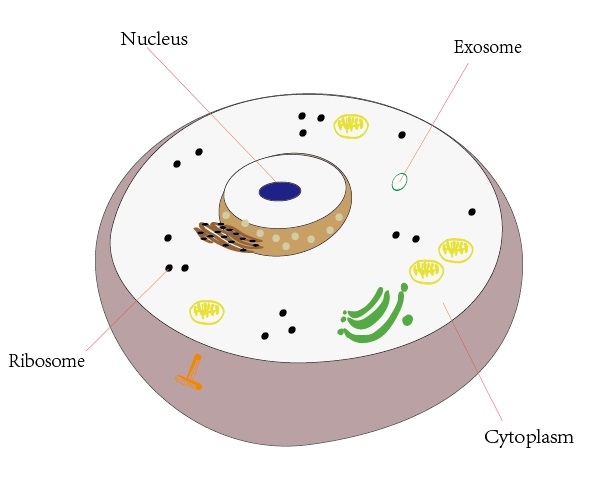
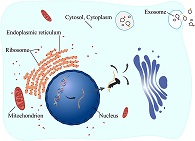
iLoc-mRNA : Predicting the subcellular location of mRNAs in human